Posts tagged science
Going for robustness: science
This is a follow-up to my earlier post entitled "Going for robustness", focusing on scientific research.
What is "robust science"? I see at least two interpretations, and I am going to discuss both of them: robustness of scientific findings, and robustness of the process of doing science, which includes in particular the robustness of the web of scientific research institutions: first and foremost universities and research labs, but also learned societies, funding agencies, publishers, etc.
Some comments on AlphaFold
Many people are asking for my opinion on the recent impressive success of AlphaFold at CASP14, perhaps incorrectly assuming that I am an expert on protein folding. I have actually never done any research in that field, but it's close enough to my research interests that I have closely followed the progress that has been made over the years. Rather than reply to everyone individually, here is a public version of my comments. They are based on the limited information on AlphaFold that is available today. I may come back to this post later and expand it.
The landscapes of digital scientific knowledge
Over the last years, an interesting metaphor for information and knowledge curation is beginning to take root. It compares knowledge to a landscape in which it identifies in particular two key elements: streams and gardens. The first use of this metaphor that I am aware of is this essay by Mike Caulfield, which I strongly recommend you to read first. In the following, I will apply this metaphor specifically to scientific knowledge and its possible evolution in the digital era.
Industrialization of scientific software: a case study
A coffee break conversion at a scientific conference last week provided an excellent illustration for the industrialization of scientific research that I wrote about in a recent blog post. It has provoked some discussion on Twitter that deserves being recorded and commented on a more permanent medium. Which is here.
The industrialization of scientific research
Over the last few years, I have spent a lot of time thinking, speaking, and discussing about the reproducibility crisis in scientific research. An obvious but hard to answer question is: Why has reproducibility become such a major problem, in so many disciplines? And why now? In this post, I will make an attempt at formulating an hypothesis: the underlying cause for the reproducibility crisis is the ongoing industrialization of scientific research.
Data science in ancient Greece
Data science is usually considered a very recent invention, made possible by electronic computing and communication technologies. Some consider it the fourth paradigm of science, suggesting that it came after three other paradigms, though the whole idea of distinct paradigms remains controversial. What I want to point out in this post is that the principles of data science are much older than most of today's practitioners imagine. Let me introduce you to Apollonius, Hipparchus, and Ptolemy, who applied these principles about 2000 years ago.
The compartmentalization of knowledge
Now that the birch pollen season is definitely over, I can draw some conclusions from a two-year experiment with the impressive sample size of one - myself. As you will see, my topic is not so much the experiment itself, but the circumstances in which it happened.
I have been allergic to birch pollen for more than thirty years. My allergy is strong enough to make normal life impossible when the birch pollen concentration is high, which happens for about three to four weeks every year. For those who have no experience with allergies, consider how sneezing five times in five minutes a few times per hour would impact your daily activities. Like most victims of pollen allergy, I consulted medical doctors in search for relief. In the course of thirty years spent in various places, even different countries, I have seen many of them, from three categories: general practitioner, otorhinolaryngologists, and allergologists. All these doctors agreed that the only reasonable treatment is antiihistamines, arguing that the only other option, immunosuppressive treatments such as cortisone, has side effects that are too severe compared to the benefit obtained.
Unfortunately, antihistamines also have a frequent side effect: drowsiness. Its degree varies between people and across different antihistamines. But in spite of undeniable progress over the years, I have yet to try an antihistamine that I could live with comfortably. I was always faced with the choice of the lesser evil: sneezing or drowsiness. I usually tried to take antihistamines as little as possible, based of birch pollen concentration forecasts, but I found that strategy hard to apply in practice.
So far for the motivation for my recent experiment. Last year I discovered, somewhat by accident, a herbalist in Paris offering a mixture of eight plant extracts for treating allergy symptoms. I asked if they considered their product sufficient as the sole treatment for a rather severe case of birch pollen allergy. They said it's worth a try, though they didn't want to make a clear promise. I tried, and it worked. Perfectly. No sneezing, no side effects. Spring 2014 was the first one I fully enjoyed since ages ago. Spring 2015 was the second. I haven't taken any antihistamines since then, nor any other allergy treatment recognized by official medecine. Of course, my new treatments has its drawbacks as well. First, it's rather expensive, about 40€ for one birch pollen season. Second, you can't take a single daily dose, you have to distribute it over the day. I followed the recommendation to dilute the daily dose in a bottle of water, which I carried with me and drank over the day.
My sample-size-one study doesn't of course permit any conclusions about the efficiency of this treatment for allergies in general, but that's not my point anyway. What I find remarkable about this story is that a small herbalist shop in Paris offers something that according to all medical doctors I ever consulted doesn't exist. Herbal remedies have been used by people all over the world for all of known history. All the eight plants in my new treatment (Plantago lanceolata, artichoke, arctium, boldo, desmodium, dandelion, horsetail, thyme) have been used by herbalists for centuries. Combining them into an efficient treatment certainly requires some solid knowledge about medical plants, but probably not a stroke of genius. How is it possible then that not even specialized allergologists are aware of such treatments? Even if it works only for 10% of pollen victims (a number I just made up), it's worth knowing about.
This compartmentalization of knowledge between traditional herbalists and 21st century medical doctors, which I suspect to be due to pure snobism, is also a lost opportunity for medical research. According to the description of my plant mixture on the Web site, its mode of action is completely different from that of antihistamines. Studying these mechanisms might well lead to new insight into the causes of pollen allergies and their treatments.
Drawing conclusions from empirical science
A recent paper in PLOS One made some noise in my twittersphere over the Christmas days. It compares the productivity of writing scientific documents using Microsoft Word and using LaTeX, and concludes that Microsoft Word is so clearly superior that, in the interest of saving taxpayers' money, scientific publishers should abandon LaTeX to allow authors to become more productive.
The noise in my twittersphere is about the technical shortcomings of the study, whose findings are in clear contradiction to the personal experience of everyone who has used both LaTeX and Microsoft Word in preparing real-life scientific articles for publication. This is well discussed in the comments on the paper. In short, the situations explored in the study are limited to the reproduction of a given piece of text with some typical "scientific" elements such as tables or formulas, but without the complexity of real-life documents: references, citations, revisions, collaborative editing, etc.
The topic of this post is a more fundamental problem illustrated by the study cited above, and which is shared by a large number of scientific explorations of much more important subjects, in particular concerning health and medicine. It is the problem of drawing practical conclusions from the results of a scientific study, such as the conclusion cited above that abandoning LaTeX would lead to significant savings in the field of scientific publishing. In the following, I will concentrate on this issue and leave aside everything else: let's assume for a few minutes that published scientific studies are 100% reliable and described clearly enough that no misunderstandings or erroneous interpretations ever occur.
The feature that the Word vs. LaTeX study shares with much of modern research is that it is purely empirical. It starts from the question if science writers are more productive using Word or using LaTeX, taking into account a few obvious parameters such as prior experience with one or the other system. To answer that question, a specific experiment is designed, performed, and analyzed. Importantly, there is no underlying model that is used to interpret the results, which is what makes the model purely empirical.
Empirical studies are characteristic of relatively young domains of scientific exploration. It's what every new field starts out with: the search for systematic relations between observable facts and quantities. As our understanding of some aspect of nature improves, we move on to the next level of scientific inquiry: the construction of models. A model makes assumptions about the mechanisms underlying the observed behavior, and allows the prediction of results that some not-yet-performed experiment should produce. The introduction of models is an enormous boost to the power and efficiency of scientific research. First of all, predictions can be tested, and therefore the models can be tested. Of course, an isolated hypothesis ("Word makes scientists more productive than LaTeX") can also be tested, but a model produces a whole family of related hypotheses that can be tested as a whole. In particular, one can search for corner cases that may be untypical from a real-world point of view, but provide a particularly precise way to test a model. Second, a model allows scientists to develop an intuitive understanding of the phenomena they are looking at, which again makes their work more efficient and more reliable. But perhaps most importantly, a model that has been exposed to several rounds of serious testing comes with a list of scenarios in which it works or doesn't work, which is a very important element in generating trust in its predictions.
As an example of a successful model, consider Newtonian mechanics as taught in high-school physics classes. It has been around for a few centuries, and its strengths and limitations are well known. Contrary to what people believed initially, it is not universally true. It breaks down for objects moving at extremely high speed, and for objects of atomic size. But it works very well for many practically relevant situations. Thanks to this and other well-tested models, engineers and architects can design engines and buildings that work as expected.
In contrast, purely empirical science provides only provisional answers to the questions asked, because it is impossible to know, or even test, that all relevant aspects of the situation have been taken into account. In the Word vs. LaTeX study, prior knowledge of either system was taken into account as a parameter, but many other factors weren't. It is conceivable, for example, that a person's native language may make them "better tuned" to one or the other system. Or their work experience, or their education. And why not genetic factors or dietary habits - this sounds far-fetched, but it can't be excluded. As long as there is no model explaining where productivity differences come from, it is not even clear what one would have to study in order to improve our understanding of the situation.
This uncertainty stemming from the existence of many unexplored potential factors makes it very risky to draw practical conclusions from purely empirical studies, no matter how well they were designed and executed. And this is a very real problem in many aspects of today's life. Suppose you are determined to adopt the "healthiest" dietary regime possible, and turn to the scientific literature for guidance. You will find a bewildering collection of partially contradicting findings. Does eating eggs expose you to a higher risk of cardiovascular diseases? Do oranges protect you against the flu? You will find studies that claim to provide the answers to such questions, but they are purely empirical and based on a small number of observations. They may even be based on experiments on mice that were extrapolated to humans. And they definitely have not explored all imaginable aspects of the question. What it vitamin C is beneficial to everyone except people with some rare blood group? What if a specific gene variant decides how your body reacts to high sugar intake? Most probably no one has ever looked into these possibilities. Not to mention the much more fundamental question if a "healthiest" diet exists at all. Perhaps the best you can do is choose between a higher risk of a stroke and a higher risk of cancer.
To end with some practical advice: the next time you see some recommendation made on a "scientific basis", check what that basis is. If it's a single recent study, it's safe to assume that the recommendation is premature. But even if it's a larger body of scientific evidence, check if there is a model behind it, and if it has been tested. If it isn't, be prepared to get a contradictory recommendation in a few years.
Reproducibility, replicability, and the two layers of computational science
The importance of reproducibility in computational science is being more and more recognized, which I think is a good sign. However, I also notice a lot of confusion about what reproducibility means exactly, and also confusion about the difference (if any) between reproducibility and replicability. I don't see a consensus yet about the exact meaning of these terms, but I would like to give my own definitions and justify them by putting them into the general context of computational science.
I'll start with the concept of reproducibility as it was used in science long before computers even existed. It refers to the reproducibility of the conclusions of a scientific study. These conclusions can take very different forms depending on the question that was being explored. It can be a simple "yes" or "no", e.g. in answering questions such as "Is the gravitational force acting in this stone the same everywhere on the Earth's surface?" or "Does ligand A bind more strongly to protein X than ligand B?" It can also be a number, as in "What is the lattice energy of NaCl?", or a mathematical function, as in "How does a spring's restoring force vary with elongation?" Any such result should come with an estimation of its precision, such as an error bar on numbers, or a reliability estimate for a yes/no answer. Reproducing a scientific conclusion means finding a "close enough" answer by performing "similar" experiments and analyses. As the terms "close enough" and "similar" show, reproducibility involves human judgement, which may well evolve over time. Reproducibility is thus not an absolute feature of a specific result, but the evaluation of a result in the context of the current state of knowledge and technology in a scientific domain. Every attempt to reproduce a given result independently (different people, tools, methods, …) augments scientific knowledge: If the reproduction leads to a "close enough" results, it provides information about the precision with which the results can be obtained, and if if doesn't, it points to some previously unrecognized crucial difference between the two experiments, which can then be explored.
Replication refers to something much more specific: repeating the exact steps in an experiment using the same (or equivalent) equipment, and comparing the outcomes. Replication is part of testing an experimental setup, or a form of quality assurance. If I measure the same quantity ten times using the same equipment and experimental samples, and get ten slightly different values, then I can use these numbers to estimate the precision of my equipment. If that precision is not sufficient for the purposes of my planned scientific study, then the equipment is not suitable.
It is useful to describe the process of doing research by a two-layer model. The fundamental layer is the technology layer: equipment and procedures that are well understood and whose precision is known from many replication attempts. On top of this, there is the research layer: the well-understood equipment is used in order to obtain new scientific information and draw conclusions from them. Any scientific project aims at improving one or the other layer, but not both at the same time. When you want to get new scientific knowledge, you use trusted equipment and procedures. When you want to improve the equipment or the procedures, you do so by doing test measurements on well-known systems. Reproducibility is a concept of the research layer, replicability belongs to the technology layer.
All this carries over identically to computational science, in principle. There is the technology layer, consisting of computers and the software that runs on them, and the research layer, which uses this technology to explore theoretical models or to interpret experimental data. Replicability belongs to the technology level. It increases trust in a computation and thus its components (hardware, software, overall workflow, provenance tracking, …). If a computation cannot be replicated, then this points to some kind of problem:
- different input data that was not recorded in the workflow (interactive user input, a random number stream initialized from the current time, …)
- a bug in the software (uninitialized variables, compiler bugs, …)
- a fault in the hardware (an unreliable memory chip, a design flaw in the processor, …)
- an ambiguous specification of the result of the computation
Ideally, the non-replicability should be eliminated, but at the very least its cause should be understood. This turns out to be very difficult in practice, in today's computing environments, essentially because case 4 is frequent and hard to avoid (today's popular programming languages are ambiguous), and because case 4 makes it impossible to identify cases 2 and 3 with certainty. I see this as a symptom of the immaturity of today's computing environments, which the computational science community should aim to improve on. The technology for removing case 4 exists. The keyword is "formal methods", and there are first attempts to apply them to scientific computing, but this remains an exotic approach for now.
As in experimental science, reproducibility belongs to the research layer and cannot be guaranteed or verified by any technology. In fact, the "reproducible research" movement is really about replicability - which is perhaps one reason for the above-mentioned confusion.
There is at the moment significant disagreement about the importance of replicability. At one end of the spectrum, there is for example Ian Gent's recomputation manifesto, which stresses the importance of replicability (which in the context of computational science he calls recomputability) because building on past work is possible only if it can be replicated as a first step. At the other end, Chris Drummond argues that replicability is "not worth having" because it doesn't contribute much to the real goal, which is reprodcucibility. It is worth reading both of these papers, because they both do a very good job at explaining their arguments. There is actually no contradiction between the two lines of arguments, the different conclusions are due to different criteria being applied: Chris Drummond sees replicability as valuable only if it improves reproducibility (which indeed it doesn't), whereas Ian Gent sees value in it for a completely different reason: it makes future research more efficient. Neither one mentions the main point in favor of replicability that I have made above: that replicability is a form of quality assurance and thus increases trust in published results.
It is probably a coincidence that both of the papers cited above use the term "computational experiment", which I think should best be avoided in this context. In the natural sciences, the term "experiment" traditionally refers to constructing a setup to observe nature, which makes experiments the ultimate source of truth in science. Computations do not have this status at all: they are applications of theoretical models, which are always imperfect. In fact, there is an interesting duality between the two: experiments are imperfect observations of the ultimate truth, whereas computations are, in the absence of buggy or ambiguous software, perfect observations of the consequences of imperfect models. Using the same term for these two concepts is a source of confusion, as I have pointed out earlier.
This fundamental difference between experiments and computations also means that replicability has a different status in experimental and computational science. When doing imperfect observations of nature, evaluating replicability is one aspect of evaluating the imperfection of the observation. Perfect observation is impossible, both due to technological limitations and for fundamental reasons (any observation modifies what is being observed). On the other hand, when computing the consequences of imperfect models, replicability does not measure the imperfections of the model, but the imperfections of the computation, which can theoretically be eliminated.
The main source of imperfections in computations is the complexity of computer software (considering the whole software stack, from the operating system to the scientific software). At this time, it is not clear if we will ever succeed in taming this complexity. Our current digital computers are chaotic systems, in which even the tiniest change (flipping a bit in memory, or replacing a single character in a program source code file) can change the result of a computation beyond any bounds. Chaotic behavior is clearly an undesirable feature in any scientific equipment (I can't think of any experimental apparatus suffering from it), but for computation we currently have no other choice. This makes quality assurance techniques, including replicability but also more standard software engineering practices such as unit testing, all the more important if we want computational results to be trustworthy.
The roles of computer programs in science
Why do people write computer programs? The answer seems obvious: in order to produce useful tools that help them (or their clients) do whatever they want to do. That answer is clearly an oversimplification. Some people write programs just for the fun of it, for example. But when we replace "people" by "scientists", and limit ourselves to the scientists' professional activities, we get a
statement that rings true: Scientists write programs because these programs do useful work for them. Lengthy computations, for example, or visualization of complex data.
This perspective of "software as a tool for doing research" is so pervasive in computational science that it is hardly ever expressed. Many scientists even see software, or perhaps the combination of computer hardware plus software as just another piece of lab equipment. A nice illustration is this TEDx lecture by Klaus Schulten about his "computational microscope", which is in fact Molecular Dynamics simulation software for studying biological macromolecules such as proteins or DNA.
To see the fallacy behind equating computer programs with lab equipment, let's take a step back and look at the basic principles of science. The ultimate goal of science is to develop an understanding of the universe that we inhabit. The specificity of science (compared to other approaches such as philosophy or religion) is that it constructs precise models for natural phenomena that it validates and improves by repeated confrontation with observations made on the real thing:
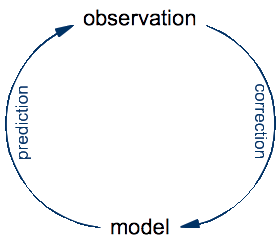
An experiment is just an optimization: it's a setup designed for making a very specific kind of observation that might be difficult or impossible to make by just looking at the world around us. The process of doing science is an eternal cycle: the model is used to make predictions of yet-to-make observations, whereas the real observations are compared to these predictions in order to validate the model and, in case of a significant discrepancies, to correct it.
In this cycle of prediction and observation, the role of a traditional microscope is to help make observations of what happens in nature. In contrast, the role of Schulten's computational microscope is to make predictions from a theoretical model. Once you think about this for a while, it seems obvious. To make observations on a protein, you need to have that protein. A real one, made of real atoms. There is no protein anywhere in a computer, so a computer cannot do observations on proteins, no matter which software is being run on it. What you look at with the computational microscope is not a protein, but a model of a protein. If you actually watch Klaus Schulten's video to the end, you will see that this distinction is made at some point, although not as clearly as I think it should be.
So it seems that the term "a tool for exploring a theoretical model" is a good description of a simulation program. And in fact that's what early simulation programs were. The direct ancestors of Schulten's computational microscope are the first Molecular Dynamics simulation programs made for atomic liquids. A classic reference is Rahman's 1964 paper on the simulation of liquid argon. The papers of that time specify the model in terms of a few mathematical equations plus a some numerical parameters. Molecular Dynamics is basically Newton's equations of motion, discretized for numerical integration, plus a simple model for the interactions between the atoms, known as the Lennard-Jones potential. A simulation program of the time was a rather straightforward translation of the equations into FORTRAN, plus some bookkeeping and I/O code. It was indeed a tool for exploring a theoretical model.
Since then, computer simulation has been applied to ever bigger and ever more complex systems. The examples shown by Klaus Schulten in his video represent the state of the art: assemblies of biological macromolecules, consisting of millions of atoms. The theoretical model for these systems is still a discretized version of Newton's equations plus a model for the interactions. But this model for the interactions has become extremely complex. So complex in fact that nobody bothers to write it down any more. It's not even clear how you would write it down, since standard mathematical notation is no longer adequate for the task. A full specification requires some algorithms and a database of chemical information. Specific aspects of model construction have been discussed at length in the scientific literature (for example how best to describe electrostatic interactions), but a complete and precise specification of the model used in a simulation-based study is never provided.
The evolution from simple simulations (liquid argon) to complex ones (assemblies of macromolecules) looks superficially like a quantitative change, but there is in fact a qualitative difference: for today's complex simulations, the computer program is the model. Questions such as "Does program X correctly implement model A?", a question that made perfect sense in the 1960s, have become meaningless. Instead, we can only ask "Does program X implement the same model as program Y?", but that question is impossible to answer in practice. The reason is that the programs are even more complex than the models, because they also deal with purely practical issues such as optimization, parallelization, I/O, etc. This phenomenon is not limited to Molecular Dynamics simulations. The transition from mathematical models to computational models, which can only be expressed in the form of computer programs, is happening in many branches of science. However, scientists are slow to recognize what is happening, and I think that is one reason for the frequent misidentification of software as experimental equipment. Once a theoretical model is complex and drowned in even more complex software, it acquires many of the characteristics of experiments. Like a sample in an experiment, it cannot be known exactly, it can only be studied by observing its behavior. Moreover, these observations are associated with systematic and statistical errors resulting from numerical issues that frequently even the program authors don't fully understand.
From my point of view (I am a theoretical physicist), this situation is not acceptable. Models play a central role in science, in particular in theoretical science. Anyone claiming to be theoretician should be able to state precisely which models he/she is using. Differences between models, and approximations to them, must be discussed in scientific studies. A prerequisite is that the models can be written down in a human-readable form. Computational models are here to stay, meaning that computer programs as models will become part of the daily bread of theoreticians. What we will have to develop is notations and techniques that permit a separation of the model aspect of a program from all the other aspects, such as optimization, parallelization, and I/O handling. I have presented some ideas for reaching this goal in this article (click here for a free copy of the issue containing it, it's on page 77), but a lot of details remain to be worked out.
The idea of programs as a notation for models is not new. It has been discussed in the context of education, for example in this paper by Gerald Sussman and Jack Wisdom, as well as in their book that presents classical mechanics in a form directly executable on a computer. The constraint of executability imposed by computer programs forces scientists to remove any ambiguities from their models. The idea is that if you can run it on your computer, it's completely specified. Sussman and Wisdom actually designed a specialized programming language for this purpose. They say it's Scheme, which is technically correct, but Scheme is a member of the Lisp family of extensible programming languages, and the extensions written by Sussman and Wisdom are highly non-trivial, to the point of including a special-purpose computer algebra system.
For the specific example that I have used above, Molecular Dynamics simulations of proteins, the model is based on classical mechanics and it should thus be possible to use the language of Sussman and Wisdom to write down a complete specification. Deriving an efficient simulation program from such a model should also be possible, but requires significant research and devlopment effort.
However, any progress in this direction can happen only when the computational science community takes a step back from its everyday occupations (producing ever more efficient tools for running ever bigger simulations on ever bigger computers) and starts thinking about the place that it occupies in the pursuit of scientific research.
Update (2014-5-26) I have also written a more detailed article on this subject.
Python as a platform for reproducible research
The other day I was looking at the release notes for the recently published release 1.8 of NumPy, the library that is the basis for most of the Scientific Python ecosystem. As usual, it contains a list of new features and improvements, but also sections such as "dropped support" (for Python 2.4 and 2.5) and "future changes", to be understood as "incompatible changes that you should start to prepare for". Dropping support for old Python releases is understandable: maintaining compatibility and testing it is work that needs to be done by someone, and manpower is notoriously scarce for projects such as NumPy. Many of the announced changes are in the same category: they permit removing old code and thus reduce maintenance effort. Other announced changes have the goal of improving the API, and I suppose they were more controversial than the others, as it is rarely obvious that one API is better than another one.
From the point of view of reproducible research, all these changes are bad news. They mean that libraries and scripts that work today will fail to work with future NumPy releases, in ways that their users, who are usually not the authors, cannot easily understand or fix. Actively maintained libraries will of course be adapted to changes in NumPy, but much, perhaps most, scientific software is not actively maintained. A PhD student doing computational reasearch might well publish his/her software along with the thesis, but then switch subjects, or leave research altogether, and never look at the old code again. There are also specialized libraries developed by small teams who don't have the resources to do as much maintenance as they would like.
Of course NumPy is not the only source of instability in the Python platform. The most visible change in the Python ecosystem is the evolution of Python itself, whose 3.x series is not compatible with the initial Python language. It is difficult to say at this time for how long Python 2.x will be maintained, but it is well possible that much of today's scientific software written in Python will become difficult to run ten years from now.
The problem of scientific publications becoming more and more difficult to use is not specific to computational science. A theoretical physicist trying to read Isaac Newton's works would have a hard time, because the mathematical language of physics has changed considerably over time. Similarly, an experimentalist trying to reproduce Galileo Galilei's experiments would find it hard to follow his descriptions. Neither is a problem in practice, because the insights obtained by Newton and Galilei have been reformulated many times since then and are available in today's language in the form of textbooks. Reading the original works is required only for studying the history of science. However, it typically takes a few decades before specific results are universally recognized as important and enter the perpetually maintained canon of science.
The crucial difference with computations is that computing platforms evolve much faster than scientific research. Researchers in fields such as physics and chemistry routinely consult original research works that are up to thirty years old. But scientific software from thirty years ago is almost certainly unusable today without changes. The state of today's software thirty years from now is likely to be worse, since software complexity has increased significantly. Thirty years ago, the only dependencies a scientific program would have is a compiler and perhaps one of a few widely known numerical libraries. Today, even a simple ten-line Python script has lots of dependencies, most of the indirectly through the Python interpreter.
One popular attitude is to say: Just run old Python packages with old versions of Python, NumPy, etc. This is an option as long as the versions you need are recent enough that they can still be built and installed on a modern computer system. And even then, the practical difficulties of working with parallel installation of multiple versions of several packages are considerable, in spite of tools designed to help with this task (have a look at EasyBuild, hashdist, conda, and Nix or its offshoot Guix).
An additional difficulty is that the installation instructions for a library or script at best mention a minimum version number for dependencies, but not the last version with which they were tested. There is a tacit assumption in the computing world that later versions of a package are compatible with earlier ones, although this is not true in practice, as the example of NumPy shows. The Python platform would be a nicer place if any backwards-incompatible change were accompanied by a change in package name. Dependencies would then be evident, and the different incompatible versions could easily be installed in parallel. Unfortunately this approach is rarely taken, a laudable exception being Pyro, whose latest incarnation is called Pyro4 to distinguish it from its not fully compatible predecessors.
I have been thinking a lot about this issue recently, because it directly impacts my ActivePapers project. ActivePapers solves the dependency versioning problem for all code that lives within the ActivePaper universe, by abandoning the notion of a single collection of "installed packages" and replacing it by explicit references to a specific published version. However, the problem persists for packages that cannot be moved inside the ActivePaper universe, typically because of extension modules written in a compiled language. The most fundamental dependencies of this kind are NumPy and h5py, which are guaranteed to be available in an ActivePapers installation. ActivePapers does record the version numbers of NumPy and h5py (and also HDF5) that were used for each individual computation, but it has currently no way to reproduce that exact environment at a later time. If anyone has a good idea for solving this problem, in a way that the average scientist can handle without becoming a professional systems administrator, please leave a comment!
As I have pointed out in an earlier post, long-term reproducibility in computational science will become possible only if the community adopts a stable code representation, which needs to be situated somewhere in between processor instruction sets and programming languages, since both ends of this spectrum are moving targets. In the meantime, we will have to live with workarounds.
A critical view of altmetrics
At first sight, altmetrics appear as an evident "update" to traditional bibliometry. It sounds pretty obvious that, as scientific communication moves on to new media and finds new forms of expressions, bibliometry should adapt. On the other hand, bibliometry is considered a more less necessary evil by most scientists. Many deplore today's "publish or perish" culture and correctly observe that it is harmful to science in the long term, giving more importance to the marketing of research studies than to their careful design and meticulous execution. I haven't yet seen any discussion of this aspect in the context of altmetrics, so I'd like to start such a discussion with this post.
First of all, why is bibliometry so popular, and why is it harmful in the long run? Second, how will this change if and when altmetrics are adopted by the scientific community?
Bibliometry provides measures of scientific activity that have two important advantages: they are objective, based on data that anyone can check in principle, and they can be evaluated by anyone, even by a computer, without any need to understand the contents of scientific papers. On the downside, those measures can only indirectly represent scientific quality precisely because they ignore the contents. Bibliometry makes the fundamental assumption that the way specific articles are received by the scientific community can be used as a proxy for quality. That assumption is, of course, wrong, and that's how bibliometry ultimately harms the progress of science.
The techniques that people use to improve their bibliometrical scores without contributing to scientific progress are well known: dilution of content (more articles with less content per article), dilution of authorship (agreements between scientists to add each others' names to their works), marketing campaigns for getting more citations, application of a single technique to lots of very similar applications even if that adds no insight whatsoever. Altmetrics will cause the same techniques to be applied to datasets and software. For example, I expect scientific software developers to take Open Source libraries and re-publish them with small modifications under a new name, in order to have their name attached to them. Unless we come up with better techniques for software installation and deployment, this will probably make the management of scientific software a bit more complicated because we will have to deal with lots of small libraries. That's a technical problem that can and should be solved with a technical solution.
However, these most direct and most discussed negative consequences of bibliometry are not the only ones and perhaps not the worst. The replacement of expert judgement by majority vote, which is the basis of bibliometry, also in its altmetrics incarnation, leads to a phenomenon which I will call "scientiic bubbles" in analogy to market bubbles in economy. A market bubble occurs if the price of a good is determined not by the people who buy it to satisfy some need, but by traders and speculators who try to estimate the future price of the good and make a profit from a rise or fall relative to the current price. In science, the "client" whose "need" is fulfilled by a scientific study is mainly future science, plus in the case of applied research engineering and product development. The role of traders and speculators is taken by referees and journal editors. A scientific bubble is a fashionable topic that many people work on not because of its scientific interest but because of the chance it provides to get a highly visible publication. Like market bubbles, scientific bubbles eventually explode when people realize that the once fashionable topic was a dead end. But before exploding, a bubble has wasted much money and intellectual energy. It may also have blocked alternative and ultimately more fruitful research projects that were refused funding because they were in contradiction with the dominating fashionable point of view.
My prediction is that altmetrics will make bubbles more numerous and more severe. One reason is the wider basis of sources from which references are counted. In today's citation-based bibliometry, citations come from articles that went through some journal's peer-reviewing process. No matter how imperfect peer review is, it does sort out most of the unfounded and obviously wrong contributions. To get a paper published in a journal whose citations count, you need a minimum of scientific competence. In contrast, anyone can publish an opinion on Twitter or Facebook. Since for any given topic the number of experts is much smaller than the number of people with just some interest, a wider basis for judgement automatically means less competence on average. As a consequence, high altmetrics scores are best obtained by writing articles that appeal to the masses who can understand what the work is about but not judge if it is well-founded. Another reason why altmetrics will contribute to bubbles is the positive feedback loop created by people reading and citing publications because they are already widely read and cited. That effect is dampened in traditional bibliometry because of the slowness of the publishing and citation mechanism.
My main argument ends here, but I will try to anticipate some criticisms and reply to them immediately.
One objection I expect is that the analysis of citation graphs can be used to assign a kind of reputation to each source and weight references by this reputation. That is the principle of Google's famous PageRank algorithm. However, any analysis of the citation graph suffers from the same fundamental problem as bibliometry itself: a method that only looks at relations between publications but not at their contents can't distinguish a gem from a shiny bubble. There will be reputation bubbles just like there are topic bubbles. No purely quantitative analysis can ever make a statement about quality. The situation is similar to mathematical formalisms, with citation graph analysis taking the role of formal proof and scientific quality the role of truth in Gödel's incompleteness theorem.
Another likely criticism is that the concept of the scientific bubble is dubious. Many paths of scientific explorations have turned out to be failures, but no one could possibly have predicted this in the beginning. In fact, many ultimately successful strategies have initially been criticized as hopeless. Moreover, exploration of a wrong path can still lead to scientific progress, once the mistake has been understood. How can one distinguish promising but ultimately wrong ideas from bubbles? The borderline is indeed fuzzy, but that doesn't mean that the concept of a bubble is useless. It's the same for market bubbles, which exist but are less severe when a good is traded both for consumption and for speculation. My point is that the bubble phenomenon exists and is detrimental to scientific progress.
Integrating scientific software and datasets into the citation record
This morning I read C. Titus Brown's blog post on how science could be so much better if scientitic data and the software used to work with it were openly available for reuse. One problem he mentions, like many others have done before, is the lack of incentive for publishing anything else but standard scientific papers. What matters for a scientist's career and for grant applications is papers, papers, papers. Any contribution that's not in a scientific journal with a reputation and an impact factor is usually ignored, even if its real impact exceeds that of many papers that nobody really wants to read.
Ideally, published scientific data and software should be treated just like a paper: it should be citeable and it should appear in the citation databases that are used to calculate impact factors, h factors, and whatever other metrics bibliometrists come up with and evaluation committees appreciate for their ease of use.
Treating text (i.e. papers), data, and code identically also happens to be useful for making scientific publications more useful to the reader, by adding interactive visualization and exploration of procedures (such as varying parameters) to the static presentation of results in a standard paper. This idea of "executable papers" has generated a lot of interest recently, as shown by Elsevier's Executable Paper Challenge and the Beyond the PDF workshop. For a technical description of how this can be achieved, see my ActivePapers project and/or the paper describing it. In the ActivePapers framework, a reference to code being called, or to a dataset being reused, is exactly identical to a reference to a published paper. It would then be much easier for citation databases to include all references rather than filter out the ones that are "classical" citations. And that's a good motivation to finally treat all scientific contributions equally.
Since the executable papers idea is much easier to sell than the idea of an upated incentive system, a seemingly innocent choice in technology could end up helping to change the way scientists and research projects are evaluated.
EuroSciPy 2011
The two keynote talks were particularly inspiring. On Saturday, Marian Petre reported on her studies of how people in general and scientists in particular develop software. The first part of her presentation was about how "expert" design and implement software, the definition of an expert being someone who produces software that actually works, is finished on time, and doesn't exceed the planned budget. The second part was about the particularities of software development in science. But perhaps the most memorable quote of the keynote was Marian's reply to a question from the audience of how to deal with unreasonable decisions coming from technically less competent managers. She recommended to learn how to manage management - a phrase that I heard repeated several times during the discussions along the conference.
The Sunday keynote was given by Fernando Perez. As was to be expected, IPython was his number one topic and there was a lot of new stuff to show off. I won't mention all the new features in the recently released version 0.11 because they are already discussed in detail elsewhere. What I find even more exciting is the new Web notebook interface, available only directly from the development site at github. A notebook is an editable trace of an interactive session that can be edited, saved, stored in a repository, or shared with others. It contains inputs and outputs of all commands. Inputs are cells that can consist of more than one line. Outputs are by default what Python prints to the terminal, but IPython provides a mechanism for displaying specific types of objects in a special way. This allows to show images (in particular plots) inline, but also to turn SymPy expressions into mathematical formulas typeset in LaTeX.
A more alarming aspect of Fernando's keynote was his statistical analysis of contributions to the major scientific libraries of the Python universe. In summary, the central packages are maintained by a grand total of about 25 people in their spare time. This observation caused a lot of debate, centered around how to encourage more people to contribute to this fundamental work.
Among the other presentations, as usual mostly of high quality, the ones that impressed me most were Andrew Straw's presentation of ROS, the Robot Operating System, Chris Myers' presentation about SloppyCell, and Yann Le Du's talk about large-scale machine learning running on a home-made GPU cluster. Not to forget the numerous posters with lots of more interesting stuff.
For the first time, EuroSciPy was complemented by domain-specific satellite meetings. I attended PyPhy, the Python in Physics meeting. Physicists are traditionally rather slow in accepting new technology, but the meeting showed that a lot of high-quality research is based on Python tools today, and that Python has also found its way into physics education at various universities.
Finally, conferences are good also because of what you learn during discussions with other participants. During EuroSciPy, I discovered a new scientific journal called Open Research Computation , which is all about software for scientific research. Scientific software developers regularly complain about the lack of visibility and recognition that their work receives by the scientific community and in particular by evaluation and grant attribution committees. A dedicated journal might just be what we need to improve the situation. I hope this will be a success.
Executable Papers
The term "executable papers" stands for the expected next revolution in scientific publishing. The move from printed journals to electronic on-line journals (or a combination of both) has changed little for authors and readers. It is the libraries that have seen the largest impact because they now do little more than paying subscription fees. Readers obtain papers as PDF files directly from the publishers' Web sites. The one change that does matter to scientists is that most journals now propose the distribute "supplementary material" in addition to the main paper. This can in principle be any kind of file, in practice it is mostly used for additional explanations, images, and tables, i.e. to keep the main paper shorter. Occasionally there are also videos, a first step towards exploring the new possibilities opened up by electronic distribution. The step to executable papers is a much bigger one: the goal is to integrate computer-readable data and executable program code together with the text part of a paper. The goals are a richer reader experience (e.g. interactive visualizations), verifiability of results by both referees and readers (by re-running part of the computations described in the paper), and re-use of data and code in later work by the same or other authors. There is some overlap in these goals with the "reproducible research" movement, whose goal is to make computational research reproducible by providing tools and methods that permit to store a trace of everything that entered into some computational procedure (input data, program code, description of the computing environment) such that someone else (or even the original author a month later) can re-run everything and obtain the same results. The new aspect in executable papers is the packaging and distribution of everything, as well as the handling of bibliographic references.
The proposals' variety mostly reflected the different background of the presenters. A mathematician documenting proofs obviously has different needs than an astrophysicist simulating a supernova on a supercomputer. Unfortunately this important aspect was never explicitly discussed. Most presenters did not even mention their field of work, much less what it implies in terms of data handling. This was probably due to the enormous time pressure; 15 to 20 minutes for a presentation plus demonstration of a complex tool was clearly not enough.
The proposals could roughly be grouped into three categories:
- Web-based tools that permit the author to compose his executable paper by supplying data, code, and text, and permit the reviewer and reader to consult this material and re-run computations.
- Systems for preserving the author's computational environment in order to permit reviewers and readers to use the author's software with little effort and without any security risks.
- Semantic markup systems that make parts of the written text interpretable by a computer for various kinds of processing
Some proposals covered two of these categories but with a clear emphasis on one of them. For the details of each propsal, see the ICCS proceedings which are freely available.
While it was interesting to see all the different ideas presented, my main impression of the Executable Paper Workshop is that of a missed opportunity. Having all those people who had thought long and hard about the various issues in one room for two days would have been a unique occasion to make progress towards better tools for the future. In fact, none of the solutions presented cover the needs of the all the domains of computational science. They make assumptions about the nature of the data and the code that are not universally valid. One or two hours of discussion might have helped a lot to improve everyone's tools.
The implementation of my own proposal, which addresses the questions of how to store code and data in a flexible, efficient, and future-proof way, is available here. It contains a multi-platform binary (MacOS, Linux, Windows, all on the x86 platform) and requires version 6 of the Java Runtime Environment. The source code is also included, but there is no build system at the moment (I use a collection of scripts that have my home-directory hard-coded in lots of places). There is, however, a tutorial. Feedback is welcome!
EuroSciPy 2010
There were lots of interesting presentations and announcements, and the breaks provided a much appreciated opportunity for exchanges between the participants. I won't try to provide an exhaustive summary, but rather list my personal highlights. Obviously this choice reflects my personal interests more than the quality of the presentations, and I will even list things that were not presented but that I learned about from other participants during the breaks.
Teaching
The opening keynote was given by Hans-Petter Langtangen, who is best known for his books about Python for scientific computing. His latest book is a textbook for a course on scientific programming for beginning science students, and the first part of his keynote was about this same course that he is teaching at the University of Oslo. As others have noted as well, he observed that the students have no problem at all with picking up Python and using it productively in science. The difficulties with using Python are elsewhere: it is hard to convince the university professors that Python is a good choice of programming language for such a course!
Another important aspect of his presentation was the observation that teaching scientific programming to beginning science students provides more than just training in some useful technique. Converting equations into programs and running them also provides a much better insight into the structure and applicability of the equations. Computational science thus helps to better educate future scientists.
Reproducible research
The reproducible research movement has the goal of improving the standards in computational science. At the moment, it is almost always impossible to reproduce published computational results from the information provided by the authors. Making these results reproducible requires a careful recording of what was calculated using which version of which software running on which machine, and of course making this information available along with the publication.
At EuroSciPy, Andrew Davison presented Sumatra, a Python library for tracking this information (and more) for computational procedures written in Python. The library is in an early stage, with more functionality to come, but those interested in reproducible research should check it out now and contribute to its development.
Jarrod Millman addressed the same topic in his presentation of the plans for creating a Foundation for Mathematical and Scientific Computing, whose goal is to fund development of tools and techniques that improve computational science.
NumPy and Python 3
As a couple of active contributors to the NumPy project were attending the conference, I asked about the state of the porting effort to Python 3. The good news is that the port is done and will soon be released. Those who have been waiting for NumPy to be ported before starting to port their own libraries can go to work right now: check out the NumPy Subversion repository, install, and use!
Useful maths libraries
Three new maths libraries that were presented caught my attention: Sebastian Walter's talk about algorithmic differentiation contained demos of algopy, a rather complete library for algorithmic differentiation in Python. During the Lightning talks on the last day, two apparently similar libraries for working with uncertain numbers (numbers with error bars) were shown: uncertainties, by Eric Lebigot, and upy, by Friedrich Romstedt. Both do error propagation and take correlations into account. Those of us working with experimental data or simulation results will appreciate this.
There was a lot more interesting stuff, of course, and I hope others will write more about it. I'll just point out that the slides for my own keynote about the future of Python in science are available from my Web site. And of course express my thanks to the organizing committee who invested a lot of effort to make this conference a big success!
Science and free will
The topic itself is an old one, perhaps as old as humanity. I won't go into its philosophical and religious aspects, but limit myself to the scientist's point of view: is free will compatible with scientific descriptions of our world? Perhaps even necessary for such descriptions? Or, on the contrary, in contradiction to the scientific approach? Can the scientific method be used to understand free will or show that it's a useless concept from the past?
What prompted me to write this post is a recent article by Anthony Cashmore in PNAS. In summary, Cashmore says that the majority of scientists do not believe in the existence of free will any more, and that society should draw conclusions from this, in particular concerning the judicial system, whose concept of responsibility for one's acts is based on a view of free will that the author no longer considers defendable. But don't take my word for it, read the article yourself. It's well written and covers many interesting points.
First of all, let me say that I don't agree at all with Cashmore's view that the judicial system should be reformed based on the prevailing view of today's scientists. I do believe that a modern society should take into account scientific findings, i.e. scientific hypotheses that have withstood a number of attempts at falsification. But mere beliefs of a small subpopulation, even if they are scientists, are not sufficient to justify a radical change of anything. As I will explain below, the question "do human beings possess free will" does not even deserve the label "scientific hypothesis" at this moment, because we have no idea of how we could answer it based on observation and experiment. We cannot claim either to be able to fully understand human behavior in terms of the laws of physics, which would allow us to call free will an unnecessary concept and invoke Occam's razor to get rid of it. Therefore, at this time, the existence of free will remains the subject of beliefs and scientists' beliefs are worth no more than anyone else's.
There is also a peculiar circularity to any argument about what "should" be done as a consequence of the non-existence of free will: if that hypothesis is true, nobody can decide anything! If humans have no free will, then societies don't have it either, and our judicial system is just as much a consequence of the laws of nature as my perceived decision to take coffee rather than tea for breakfast this morning.
Back to the main topic of this post: the relation between science and free will. It starts with the observation of a clear conflict. Science is about identifying regularities in the world that surrounds us, which permit the construction of detailed and testable theories. The first scientific theories were all about deterministic phenomena: given the initial state of some well-defined physical system (think of a clockwork, for example), the state of the system at any time in the future can be predicted with certainty. Later, stochastic phenomena entered the scientific world view. With stochasticity, the detailed behavior of a system is no longer predictable, but certain average properties still are. For example, we can predict how the temperature and pressure of water will change when we heat it, even though we cannot predict how each individual molecule will move. It is still a subject of debate whether stochastic elements exist in the fundamental laws of nature (quantum physics being the most popular candidate), or if they are merely a way of describing complex systems whose state we cannot analyze in detail due to insufficient resources. But scientists agree that a scientific theory may contain two forms of causality: determinism and stochasticity.
Free will, if it exists, would have to be added as a third form of causality. But it is hard to see how this could be done. The scientific method is based on identifying conditions from which exact predictions can be made. The decisions of an agent that possesses free will are by definition unpredictable, and therefore any theory about a system containing such an agent would be impossible to verify. Therefore the scientific method as we know it today cannot possibly take into consideration the existence of free will. Obviously this makes it impossible to examine the existence of free will as a scientific hypothesis. It also means that a hard-core scientist, who considers the scientific method as the only way to establish truth, has to deny the existence of free will, or else accept that some important aspects of our universe are forever inaccessible to scientific investigation.
However, there is another aspect to the relation between science and free will, which I haven't seen discussed yet anywhere: the existence of free will is in fact a requirement for the scientific method! Not as part of a system under scientific scrutiny, but as part of the scientist who runs an investigation. Testing a scientific hypothesis requires at the very least observing a specific phenomenon, but in most cases also preparing a well-defined initial state for some system that will then become the subject of observation. A scientist decides to create an experimental setup to verify some hypothesis. If the scientist were just a complex machine whose behavior is governed by the very same laws that he believes to be studying, then his carefully thought-out experiment is nothing but a particularly probable outcome of the laws of nature. We could still draw conclusions from observing it, of course, but these observations then only provide anecdotical evidence that is no more relevant than what we get from passively watching things happen around us.
In summary, our current scientific method supposes the existence of free will as an attribute of scientists, but also its absence from any system subjected to scientific scrutiny. This poses limits to what scientific investigation can yield when applied to humans.
Scientific computing needs deterministic programming paradigms
A programming paradigm defines a general approach to writing programs. It defines the concepts and abstractions in terms of which a program is expressed. For example, a programming paradigm defines how data is described, how control flow is handled, how program elements are composed, etc. Well-known programming paradigms are structured programming, object-oriented programming, and functional programming.
The implementation of a programming paradigm consists of a programming language, its runtime system, libraries, and sometimes coding conventions. Some programming languages are optimized for a specific paradigm, whereas others are explicitly designed to support multiple paradigms. Paradigms that the language designer did not have in mind can sometimes be implemented by additional conventions, libraries, or preprocessors.
The list of programming paradigms that have been proposed and/or used is already quite long (see the Wikipedia entry, for example), but the ones that are practically important and significantly distinct are much less numerous. A good overview and comparison is given in the book chapter "Programming paradigms for dummies" by Peter van Roy. I will concentrate on one aspect discussed in van Roy's text (look at section 6 in particular), which I consider of particular relevance for scientific computing: determinism.
A deterministic programming paradigm is one in which every possible program has a fully deterministic behaviour: given the same input, it executes its steps in the same order and produces the same output. This is in fact what most of us would intuitively expect from a computer program. However, there are useful programs that could not be written with this restriction. A Web server, for example, has to react to external requests which are outside of its control, and take into account resource usage (e.g. database access) and possible network errors in deciding when and in which order to process requests. This shows that there is a need for non-deterministic programming paradigms. For the vast majority of scientific applications, however, determinism is a requirement, and a programming paradigm that enforces determinism is a big help in avoiding bugs. Most scientific applications that run serially have been written using a deterministic programming paradigm, as implemented by most of the popular programming languages.
Parallel computing has changed the situation significantly. When several independent processors work together on the execution of an algorithm, fully deterministic behavior is no longer desirable, as it would imply frequent synchronizations of all processors. The precise order in which independent operations are executed is typically left unspecified by a program. What matters is that the output of the program is determined only by the input. As long as this is guaranteed, it is acceptable and even desirable to let compilers and run-time systems optimize the scheduling of individual subtasks. In Peter van Roy's classification, this distinction is called "observable" vs. "non-observable" non-determinism. A programming paradigm for scientific computing should permit non-determinism, but should exclude observable non-determinism. While observable non-determinism makes the implementation of certain programs (such as Web servers) possible, it also opens the way to bugs that are particularly nasty to track down: deadlocks, race conditions, results that change with the number of processors or when moving from one parallel machine to another one.
Unfortunately, two of the most popular programming paradigms for parallel scientific applications do allow observable non-determinism: message passing, as implemented by the MPI library, and multi-threading. Those who have used either one have probably suffered the consequences. The problem is thus well known, but the solutions aren't. Fortunately, they do exist: there are several programming paradigms that encapsulate non-determinism in such a way that it cannot influence the results of a program. One of them is widely known and used: OpenMP, which is a layer above multi-threading that guarantees deterministic results. However, OpenMP is limited to shared-memory multiprocessor machines.
For the at least as important category of distributed-memory parallel machines, there are also programming paradigms that don't have the non-deterministic features of message passing, and they are typically implemented as a layer above MPI. One example is the BSP model, which I have presented in an article in the magazine Computing in Science and Engineering. Another example is the parallel skeletons model, presented by Joël Falcou in the same magazine. Unfortunately, these paradigms are little known and not well supported by programming tools. As a consequence, most scientific applications for distributed-memory machines are written using the message passing paradigm.
Finally, a pair of programming paradigms discussed by van Roy deserves special mention, because it might well become important in scientific computing in the near future: functional programming and declarative concurrency. I have written about functional programming earlier; its main advantage is the possibility to apply mathematical reasoning and automatic transformations to program code, leading to better code (in the sense of correctness) and to better optimization techniques. Declarative concurrency is functional programming plus annotations for parallelization. The nice feature is that these annotations (not very different in principle from OpenMP pragmas) don't change the result of the program, they only influence its performance on a parallel machine. Starting from a correct functional program, it is thus possible to obtain an equivalent parallel one by automatic or manual (but automatically verified) transformations that is guaranteed to behave identically except for performance. Correctness and performance can thus be separated, which should be a big help in writing correct and efficient parallel programs. I say "should" because this approach hasn't been used very much, and isn't supported yet by any mainstream programming tools. This may change in a couple of years, so you might want to watch out for developments in this area.
Sheldrake's New Science of Life
The question that Sheldrake addresses in this book is where form comes from. What defines the arrangements of atoms in a molecule? Or in a crystal? Why do proteins fold into their characteristic structures? How do biological molecules assemble into cells? And how to cells divide and specialize to form an embryo?
The standard reply to these questions you can find in science textbooks is that all these forms come from the fundamental interactions of physics. Molecules are simply energetically favorable arrangements of atoms. Proteins fold in such a way that free energy is minimized. Cells assemble as a result of complex attractive interactions between its constituents, which ultimately can be reduced to fundamental physics. Embryos develop according to a "genetic program" stored in the fecundated egg's DNA.
As Sheldrake rightly emphasizes, even though it may come as a surprise to most non-experts, these affirmations cannot be verified. They express a common belief among practicing scientists, and they are compatible with everything we know about nature, but they may well be wrong. We simply cannot verify them because the fundamental equations of physics can be solved only for very simple systems. Even for one of the simplest molecules, water, we cannot predict the arrangement of its atoms directly from the basic principles of physics. What we use in practice are approximations, but these approximations have been selected because they permit to predict the known molecular structures. We cannot use such approximations to verify more fundamental problems.
Sheldrake proposes an alternative theory, based on what he calls "morphogenetic fields". From my point of view as a physicist, the name is not very well chosen because these entities do not correspond to what a physicist would call a field, but of course this term may be perfectly clear to biologists. It's a minor point because Sheldrake explains this concept very clearly in his book. In summary, his theory says that forms exist because they have existed before; atoms, molecules, and cells arrange themselves into patterns that they "remember" from the past. His morphogenetic fields are a giant database of forms that the universe keeps around forever.
The main "problem" with this theory is that if it is right, even just approximately, then standard science, from physics to biology, is very much wrong. It is probably for this reason that his book has attracted so much criticism from the science establishment. Otherwise, there is little one could criticize: Sheldrake explains his theory and its consequences for chemistry and biology, and he proposes a large number of experimental verifications that would permit to test it. This is science at its best. Of course his theory may turn out to require modifications, or even be completely wrong, but that is true of any scientific theory when it is first formulated.
In fact, I recommend this book to anyone interested in the scientific process because of its detailed discussion of how scientific discovery works. I haven't seen many books accessible to non-specialists that explain the limits of verifiability of a scientific theory, for example. Nor have I seen any other book that makes the distinction between verified theories and widely accepted but untested beliefs so clear as Sheldrake does. Even if you don't care about his theory, you can gain a lot from reading this book.
Tags: computational science, computer-aided research, emacs, mmtk, mobile computing, polycrisis, programming, proteins, python, rants, reproducible research, science, scientific computing, scientific software, social networks, software, source code repositories, sustainable software
By month: 2026-03, 2025-11, 2025-06, 2025-04, 2025-03, 2024-10, 2023-11, 2023-10, 2022-08, 2021-06, 2021-01, 2020-12, 2020-11, 2020-07, 2020-05, 2020-04, 2020-02, 2019-12, 2019-11, 2019-10, 2019-05, 2019-04, 2019-02, 2018-12, 2018-10, 2018-07, 2018-05, 2018-04, 2018-03, 2017-12, 2017-11, 2017-09, 2017-05, 2017-04, 2017-01, 2016-05, 2016-03, 2016-01, 2015-12, 2015-11, 2015-09, 2015-07, 2015-06, 2015-04, 2015-01, 2014-12, 2014-09, 2014-08, 2014-07, 2014-05, 2014-01, 2013-11, 2013-09, 2013-08, 2013-06, 2013-05, 2013-04, 2012-11, 2012-09, 2012-05, 2012-04, 2012-03, 2012-02, 2011-11, 2011-08, 2011-06, 2011-05, 2011-01, 2010-07, 2010-01, 2009-09, 2009-08, 2009-06, 2009-05, 2009-04