Recent posts
A first experience with Open Access publishing
Most scientists have found out by now that a lot has been going wrong with scientific publishing over the years. In many fields, scientific journals are no longer fulfilling what used to be their primary role: disseminating and archiving the results of scientific studies. One of the new approaches that were developed to fix the publishing system is Open Access: the principle that published articles should be freely accessible to everyone (under conditions that vary according to which "dialect" of Open Access is used) and that the cost of the publishing procedure should be payed in some other way than subscription fees. The universe of Open Access publishing has become quite complex in itself. For those who want to know more about it, a good starting point is this book, whose electronic form is, of course, Open Access.
While I have been following the developments in Open Access publishing for a few years, I had never published any Open Access article myself. I work at the borderline of theoretical physics and biophysics, which sounds like closely related fields but they nevertheless have very different publishing traditions. In theoretical physics, the most well-known journals are produced by non-commercial publishers, in particular scientific societies. Their prices have not exploded, nor do these publishers put pressure on libraries to subscribe to more than they want to. There is a also a strong tradition of making preprints freely available, e.g. on arXiv.org. This combined model continues to work well for theoretical physics, meaning that there is little incentive to look at Open Access publishing models. However, as soon as the "bio" prefix comes into play, the main journals are commercial. Some offer a per-article Open Access option, in exchange for the authors paying a few hundred to a few thousand dollars per article. There are also pure Open Access journals covering this field (e.g. PLOS Computational Biology), whose price range is similar. On the scale of the working budget of a theoretician working in France, these publishing fees are way too high, which is why I never considered Open Access for my "applied" research.
The fact that I have recently published my first Open Access article, in the pure Open Access journal F1000Research, is almost a bit accidental. The topic of the article is the role of computation in science, with a particular emphasis on the necessity to keep scientific models distinct from software tools. I had the plan to write such an artile for a while, but it didn't really fit into any of the journals I knew. The subject is computational science, but more its philosophical foundations than the technicalities that journals on computational science specialize in. The audience is scientists applying computations, which is a much larger group than the methodology specialists who subscribe to and read computational science journals. Even if some computational science journal might have accepted my article, it wouldn't have reached most of its intended audience. A journal on the philosphy of science would have been worse, as almost no practitioner of computational science looks at this literature. Since there was no clear venue where the intended audience would have a chance of finding my article, the best option was some Open Access journal where at least the article would be accessible to everyone. Publicity through social networks could then help potentially interested readers discover it. Two obstacles remained: finding an Open Access journal with a suitable subject domain, and getting around the money problem.
At the January 2014 Community Call of the Mozilla Science Lab, I learned that F1000Research was starting a new section on "science communication", and was waiving article processing charges for that section in 2014. This was confirmed shortly thereafter on the journal's blog. Science communication was in fact a very good label for what I wanted to write about. And F1000Research looked like an interesting journal to test because its attitude to openness goes beyond Open Access: the review process is open as well, meaning that reviews are published with the reviewers' names, and get their own DOI for reference. So there was my opportunity.
For those new to the Open Access world, I will give a quick overview of the submission and publishing process. Everything is handled online, through the journal's Web site and by e-mail. Since I very much prefer writing LaTeX to using Word, I chose the option of submitting through the writeLaTeX service. The idea of writeLaTeX is that you edit your article using their Web tools, but nothing stops you from downloading the template provided by F1000Research, writing locally, and uploading the final text in the end. I thus wrote my article using my preferred tool (Emacs) and on my laptop even when I didn't have a network connection. Once you submit your article, it is revised by the editorial staff (concerning language, style, and layout, they don't touch the contents). Once you approve the revision, the article is published almost instantaneously on the journal Web site. You are then asked to suggest reviewers, and the journal asks some of them (I don't know how they make their choice) to review the article. Reviews are published as they come in, and you get an e-mail alert. In addition to providing detailed comments, reviewers judge the article as "approved", "approved with reservations" or "not approved". As soon as two reviewers "approve", the article status changes to "indexed", meaning that it gets a DOI and it is listed in databases such as PubMed or Scopus. Authors can reply to reviewers (again in public), and they are encouraged to revise their article based on the reviewers' suggestions. All versions of an article remain accesible indefinitely on the journal's Web site, so the history of the article remains accessible forever.
Overall I would judge my experience with F1000Research as very positive. The editorial staff replies rapidly and gets problems solved (in my case, technical problems with the Web site). Open review is much more reasonable than the traditional secret peer review process. No more guessing who the reviewers are in order to please them with citations with the hope of getting your revision accepted rapidly. No more lengthy letters to the editor trying to explain diplomatically that the reviewer is incompetent. With open reviewing, authors and reviewers act as equals, as it should always have been.
The only criticism I have concerns a technical point that I hope will be improved in the future. Even if you submit your original article through writeLaTeX, you have to prepapre revisions using Microsoft Word: you download a Word file for the initially published version, activate "track changes" mode, make your changes, and send the file back. For someone who doesn't have Microsoft Word, or is not familiar with its operation, this is an enormous barrier. A journal that encourages authors to revise their articles should also allow them to do so using tools that they have and are familiar with.
Will I publish in F1000Research again? I don't expect to do so in the near future. With the exception of the science communication section, F1000Research is heavily oriented towards the life sciences, so most of my research doesn't fit in. And then there is the money problem. Without the waiver mentioned above, I'd have had to pay 500 USD for my manuscript classified as an "opinion article". Regular research articles are twice as much. Compared to a theoretician's budget, which needs to cover mostly travel, these amounts are important. Moreover, in France's heavily bureaucratized public research, every euro comes with strings attached that define when, where, and on what you are allowed to spend it. Project-specific research grants often do allow to pay publication costs, but research outside of such projects, which is still common in the theoretical sciences, doesn't have any specific budget to turn to. The idea of the Open Access movement is to re-orient the money currently spent on subscriptions towards paying publishing costs directly, but such decisions are made on a political and administrational level very remote from my daily work. Until they happen, it is rather unlikely that I will publish in Open Access mode again.
Exploring Racket
Over the last few months I have been exploring the Racket language for its potential as a language for computational science, and it's time to summarize my first impressions.
Why Racket?
There are essentially two reasons for learning a programing language: (1) getting acquainted with a new tool that promises to get some job done better than with other tools, and (2) learning about other approaches to computing and programming. My interest in Racket was driven by a combination of these two aspects. My background is in computational science (phsyics, chemistry, and structural biology), so I use computation extensively in my work. Like most computational scientists of my generation, I started working in Fortran, but quickly found this unsatisfactory. Looking for a better way to do computational science, I discovered Python in 1994 and joined the Matrix-SIG that developed what is now known as NumPy. Since then, Python has become my main programming language, and the ecosystem for scientific computing in Python has flourished to a degree unimaginable twenty years ago. For doing computational science, Python is one of the top choices today.
However, we shouldn't forget that we are still living in the stone age of computational science. Fortran was the Paleolithic, Python is the Neolithic, but we have to move on. I am convinced that computing will become as much an integral part of doing science as mathematics, but we are not there yet. One important aspect has not evolved since the beginnings of scientific computing in the 1950s: the work of a computational scientist is dominated by the technicalities of computing, rather than by the scientific concerns. We write, debug, optimize, and extend software, port it to new machines and operating systems, install messy software stacks, convert file formats, etc. These technical aspects, which are mostly unrelated to doing science, take so much of our time and attention that we think less and less about why we do a specific computation, how it fits into more general theoretical frameworks, how we can verify its soundness, and how we can improve the scientific models that underly our computations. Compare this to how theoreticians in a field like physics or chemistry use mathematics: they have acquired most of their knowledge and expertise in mathematics during their studies, and spend much more time applying mathematics to do science than worrying about the intrinsic problems of mathematics. Computing should one day have the same role. For a more detailed description of what I am aiming at, see my recent article.
This lengthy foreword was necessary to explain what I am looking for in Racket: not so much another language for doing today's computational science (Python is a better choice for that, if only for its well-developed ecosystem), but as an evironment for developing tomorrow's computational science. The Racket Web site opens with the title "A programmable programming language", and that is exactly the aspect of Racket that I am most interested in.
There are two more features of Racket that I found particularly attractive. First, it is one of the few languages that have good support for immutable data structures without being extremist about it. Mutable state is the most important cause of bugs in my experience (see my article on "Managing State" for details), and I fully agree with Clojure's Rich Hickey who says that "immutability is the right default". Racket has all the basic data structures in a mutable and an immutable variant, which provides a nice environment to try "going immutable" in practice. Second, there is a statically typed dialect called Typed Racket which promises a straightforward transition from fast prototyping in plain Racket to type-safe and more efficient production code in Typed Racket. I haven't looked at this yet, so I won't say any more about it.
Racket characteristics
For readers unfamiliar with Racket, I'll give a quick overview of the language. It's part of the Lisp family, more precisely a derivative of Scheme. In fact, Racket was formerly known as "PLT Scheme", but its authors decided that it had diverged sufficiently from Scheme to give it a different name. People familiar with Scheme will still recognize much of the language, but some changes are quite profound, such as the fact that lists are immutable. There are also many extensions not found in standard Scheme implementations.
The hallmark of the Lisp family is that programs are defined in terms of data structures rather than in terms of a text-based syntax. The most visible consequence is a rather peculiar visual aspect, which is dominated by parentheses. The more profound implication, and in fact the motivation for this uncommon choice, is the equivalence of code and data. Program execution in Lisp is nothing but interpretation of a data structure. It is possible, and common practice, to construct data structures programmatically and then evaluate them. The most frequent use of this characteristic is writing macros (which can be seen as code preprocessors) to effectively extend the language with new features. In that sense, all members of the Lisp family are "programmable programming languages".
However, Racket takes this approach to another level. Whereas traditional Lisp macros are small code preprocessors, Racket's macro system feels more like a programming API for the compiler. In fact, much of Racket is implemented in terms of Racket macros. Racket also provides a way to define a complete new language in terms of existing bits and pieces (see the paper "Languages as libraries" for an in-depth discussion of this philosophy). Racket can be seen as a construction kit for languages that are by design interoperable, making it feasible to define highly specific languages for some application domain and yet use it in combination with a general-purpose language.
Another particularity of Racket is its origin: it is developed by a network of academic research groups, who use it as tool for their own research (much of which is related to programming languages), and as a medium for teaching. However, contrary to most programming languages developed in the academic world, Racket is developed for use in the "real world" as well. There is documentation, learning aids, development tools, and the members of the core development team are always ready to answer questions on the Racket user mailing list. This mixed academic-application strategy is of interest for both sides: researchers get feedback on the utility of their ideas and developments, and application programmers get quick access to new technology. I am aware of only three other languages developed in a similar context: OCaml, Haskell, and Scala.
Learning and using Racket
A first look at the Racket Guide (an extended tutorial) and the Racket Reference shows that Racket is not a small language: there is a bewildering variety of data types, control structures, abstraction techniques, program structuration methods, and so on. Racket is a very comprehensive language that allows both fine-tuning and large-scale composition. It definitely doesn't fit into the popular "low-level" vs. "high-level" dichotomy. For the experienced programmer, this is good news: whatever technique you know to be good for the task at hand is probably supported by Racket. For students of software development, it's probably easy to get lost. Racket comes with several subsets developed for pedagogical purposes, which are used in courses and textbooks, but I didn't look at those. What I describe here is the "standard" Racket language.
Racket comes with its own development environment called "DrRacket". It looks quite poweful, but I won't say more about it because I haven't used it much. I use too many languages to be interested in any language-specific environment. Instead, I use Emacs for everything, with Geiser for Racket development.
The documentation is complete, precise, and well presented, including a pleasant visual layout. But it is not always an easy read. Be prepared to read through some background material before understanding all the details in the reference documentation of some function you are interested in. It can be frustrating sometimes, but I have never been disappointed: you do find everything you need to know if you just keep on following links.
My personal project for learning Racket is an implementation of the MOSAIC data model for molecular simulations. While my implementation is not yet complete (it supports only two kinds of data items, universes and configurations), it has data structure definitions, I/O to and from XML, data validation code, and contains a test suite for everything. It uses some advanced Racket features such as generators and interfaces, not so much out of necessity but because I wanted to play with them.
Overall I had few surprises during my first Racket project. As I already said, finding what you need in the documentation takes a lot of time initially, mostly because there is so much to look at. But once you find the construct you are looking for, it does what you expect and often more. I remember only one ongoing source of frustration: the multitude of specialized data structures, which force you to make choices you often don't really care about, and to insert conversion functions when function A returns a data structure that isn't exactly the one that function B expects to get. As an illustration, consider the Racket equivalent of Python dictionaries, hash tables. They come in a mutable and an immutable variant, each of which can use one of three different equality tests. It's certainly nice to have that flexibility when you need it, but when you don't, you don't want to have to read about all those details either.
As for Racket's warts, I ran into two of them. First, the worst supported data structure in Racket must be the immutable vector, which is so frustrating to work with (every operation on an immutable vector returns a mutable vector, which has to be manually converted back to an immutable vector) that I ended up switching to lists instead, which are immutable by default. Second, the distinction (and obligatory conversion) between lists, streams, generators and a somewhat unclear sequence abstraction makes you long for the simplicity of a single sequence interface as found in Python or Clojure. In Racket, you can decompose a list into head and tail using first and rest. The same operations on a stream are stream-first and stream-rest. The sequence abstraction, which covers both lists and streams and more, has sequence-tail for the tail, but to the best of my knowledge nothing for getting the first element, other than the somewhat heavy (for/first ([element sequence]) element).
The macro requirements of my first project were modest, not exceeding what any competent Lisp programmer would easily do using defmacro (which, BTW, exists in Racket for compatibility even though its use is discouraged). Nevertheless, in the spirit of my exploration, I tried all three levels of Racket's hygienic macro definitions: syntax-rule, syntax-case, and syntax-parse, in order of increasing power and complexity. The first, syntax-rule is straightforward but limited. The last one, syntax-parse, is the one you want for implementing industrial-strength compiler extensions. I don't quite see the need for the middle one, syntax-case, so I suppose it's there for historical reasons, being older than syntax-parse. Macros are the one aspect of Racket for which I recommend starting with something else than the Racket documentation: Greg Hendershott's Fear of Macros is a much more accessible introduction.
Scientific computing
As I said in the beginning of this post, my goal in exploring Racket was not to use it for my day-to-day work in computational science, but nevertheless I had a look at the support for scientific computing that Racket offers. In summary, there isn't much, but what there is looks very good.
The basic Racket language has good support for numerical computation, much of which is inherited from Scheme. There are integers of arbitrary size, rational numbers, and floating-point numbers (single and double precision), all with the usual operations. There are also complex numbers whose real/imaginary parts can be exact (integer or rational) or inexact (floats). Unlimited-precision floats are provided by an interface to MPFR in the Racket math library.
The math library (which is part of every standard Racket installation) offers many more goodies: multidimensional arrays, linear algebra, Fourier transforms, special functions, probability distributions, statistics, etc. The plot library, also in the standard Racket installation, adds one of the nicest collections of plotting and visualization routines that I have seen in any language. If you use DrRacket, you can even rotate 3D scenes interactively, a feature that I found quite useful when I used (abused?) plots for molecular visualization.
Outside of the Racket distribution, the only library I could find for scientific applications is Doug Williams' "science collection", which predates the Racket math library. It looks quite good as well, but I didn't find an occasion yet for using it.
Could I do my current day-to-day computations with Racket? A better way to put it is, how much support code would I have to write that is readily available for more mature scientific languages such as Python? What I miss most is access to my data in HDF5 and netCDF formats. And the domain-specific code for molecular simulation, i.e. the equivalent of my own Molecular Modeling Toolkit. Porting the latter to Racket would be doable (I wrote it myself, so I am familiar with all the algorithms and its pitfalls), and would in fact be an opportunity to improve many details. But interfacing HDF5 or netCDF sounds like a lot of work with no intrinsic interest, at least to me.
The community
Racket has an apparently small but active, competent, and friendly community. I say "apparently" because all I have to base my judgement on is the Racket user mailing list. Given Racket's academic and teaching background, it is quite possible that there are lots of students using Racket who find sufficient support locally that they never manifest themselves on the mailing list. Asking a question on the mailing list almost certainly leads to a competent answer, sometimes from one of the core developers, many of whom are very present. There are clearly many Racket beginners (and also programming newbies) on the list, but compared to other programming language users' lists, there are very few naive questions and comments. It seems like people who get into Racket are serious about programming and are aware that problems they encounter are most probably due to their lack of experience rathen than caused by bugs or bad design in Racket.
I also noticed that the Racket community is mostly localized in North America, judging from the peak posting times on the mailing list. This looks strange in today's Internet-dominated world, but perhaps real-life ties still matter more than we think.
Even though the Racket community looks small compared to other languages I have used, it is big and healthy enough to ensure its existence for many years to come. Racket is not the kind of experimental language that is likely to disappear when its inventor moves on to the next project.
Conclusion
Overall I am quite happy with Racket as a development language, though I have to add that I haven't used it for anything mission-critical yet. I plan to continue improving and completing my Racket implementation of Mosaic, and move it to Typed Racket as much as possible. But I am not ready to abandon Python as my workhorse for computational science, there are simply too many good libraries in the scientific Python ecosystem that are important for working efficiently.
The roles of computer programs in science
Why do people write computer programs? The answer seems obvious: in order to produce useful tools that help them (or their clients) do whatever they want to do. That answer is clearly an oversimplification. Some people write programs just for the fun of it, for example. But when we replace "people" by "scientists", and limit ourselves to the scientists' professional activities, we get a
statement that rings true: Scientists write programs because these programs do useful work for them. Lengthy computations, for example, or visualization of complex data.
This perspective of "software as a tool for doing research" is so pervasive in computational science that it is hardly ever expressed. Many scientists even see software, or perhaps the combination of computer hardware plus software as just another piece of lab equipment. A nice illustration is this TEDx lecture by Klaus Schulten about his "computational microscope", which is in fact Molecular Dynamics simulation software for studying biological macromolecules such as proteins or DNA.
To see the fallacy behind equating computer programs with lab equipment, let's take a step back and look at the basic principles of science. The ultimate goal of science is to develop an understanding of the universe that we inhabit. The specificity of science (compared to other approaches such as philosophy or religion) is that it constructs precise models for natural phenomena that it validates and improves by repeated confrontation with observations made on the real thing:
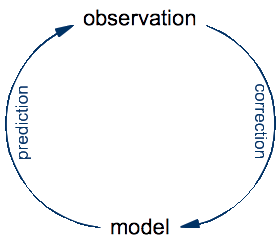
An experiment is just an optimization: it's a setup designed for making a very specific kind of observation that might be difficult or impossible to make by just looking at the world around us. The process of doing science is an eternal cycle: the model is used to make predictions of yet-to-make observations, whereas the real observations are compared to these predictions in order to validate the model and, in case of a significant discrepancies, to correct it.
In this cycle of prediction and observation, the role of a traditional microscope is to help make observations of what happens in nature. In contrast, the role of Schulten's computational microscope is to make predictions from a theoretical model. Once you think about this for a while, it seems obvious. To make observations on a protein, you need to have that protein. A real one, made of real atoms. There is no protein anywhere in a computer, so a computer cannot do observations on proteins, no matter which software is being run on it. What you look at with the computational microscope is not a protein, but a model of a protein. If you actually watch Klaus Schulten's video to the end, you will see that this distinction is made at some point, although not as clearly as I think it should be.
So it seems that the term "a tool for exploring a theoretical model" is a good description of a simulation program. And in fact that's what early simulation programs were. The direct ancestors of Schulten's computational microscope are the first Molecular Dynamics simulation programs made for atomic liquids. A classic reference is Rahman's 1964 paper on the simulation of liquid argon. The papers of that time specify the model in terms of a few mathematical equations plus a some numerical parameters. Molecular Dynamics is basically Newton's equations of motion, discretized for numerical integration, plus a simple model for the interactions between the atoms, known as the Lennard-Jones potential. A simulation program of the time was a rather straightforward translation of the equations into FORTRAN, plus some bookkeeping and I/O code. It was indeed a tool for exploring a theoretical model.
Since then, computer simulation has been applied to ever bigger and ever more complex systems. The examples shown by Klaus Schulten in his video represent the state of the art: assemblies of biological macromolecules, consisting of millions of atoms. The theoretical model for these systems is still a discretized version of Newton's equations plus a model for the interactions. But this model for the interactions has become extremely complex. So complex in fact that nobody bothers to write it down any more. It's not even clear how you would write it down, since standard mathematical notation is no longer adequate for the task. A full specification requires some algorithms and a database of chemical information. Specific aspects of model construction have been discussed at length in the scientific literature (for example how best to describe electrostatic interactions), but a complete and precise specification of the model used in a simulation-based study is never provided.
The evolution from simple simulations (liquid argon) to complex ones (assemblies of macromolecules) looks superficially like a quantitative change, but there is in fact a qualitative difference: for today's complex simulations, the computer program is the model. Questions such as "Does program X correctly implement model A?", a question that made perfect sense in the 1960s, have become meaningless. Instead, we can only ask "Does program X implement the same model as program Y?", but that question is impossible to answer in practice. The reason is that the programs are even more complex than the models, because they also deal with purely practical issues such as optimization, parallelization, I/O, etc. This phenomenon is not limited to Molecular Dynamics simulations. The transition from mathematical models to computational models, which can only be expressed in the form of computer programs, is happening in many branches of science. However, scientists are slow to recognize what is happening, and I think that is one reason for the frequent misidentification of software as experimental equipment. Once a theoretical model is complex and drowned in even more complex software, it acquires many of the characteristics of experiments. Like a sample in an experiment, it cannot be known exactly, it can only be studied by observing its behavior. Moreover, these observations are associated with systematic and statistical errors resulting from numerical issues that frequently even the program authors don't fully understand.
From my point of view (I am a theoretical physicist), this situation is not acceptable. Models play a central role in science, in particular in theoretical science. Anyone claiming to be theoretician should be able to state precisely which models he/she is using. Differences between models, and approximations to them, must be discussed in scientific studies. A prerequisite is that the models can be written down in a human-readable form. Computational models are here to stay, meaning that computer programs as models will become part of the daily bread of theoreticians. What we will have to develop is notations and techniques that permit a separation of the model aspect of a program from all the other aspects, such as optimization, parallelization, and I/O handling. I have presented some ideas for reaching this goal in this article (click here for a free copy of the issue containing it, it's on page 77), but a lot of details remain to be worked out.
The idea of programs as a notation for models is not new. It has been discussed in the context of education, for example in this paper by Gerald Sussman and Jack Wisdom, as well as in their book that presents classical mechanics in a form directly executable on a computer. The constraint of executability imposed by computer programs forces scientists to remove any ambiguities from their models. The idea is that if you can run it on your computer, it's completely specified. Sussman and Wisdom actually designed a specialized programming language for this purpose. They say it's Scheme, which is technically correct, but Scheme is a member of the Lisp family of extensible programming languages, and the extensions written by Sussman and Wisdom are highly non-trivial, to the point of including a special-purpose computer algebra system.
For the specific example that I have used above, Molecular Dynamics simulations of proteins, the model is based on classical mechanics and it should thus be possible to use the language of Sussman and Wisdom to write down a complete specification. Deriving an efficient simulation program from such a model should also be possible, but requires significant research and devlopment effort.
However, any progress in this direction can happen only when the computational science community takes a step back from its everyday occupations (producing ever more efficient tools for running ever bigger simulations on ever bigger computers) and starts thinking about the place that it occupies in the pursuit of scientific research.
Update (2014-5-26) I have also written a more detailed article on this subject.
Python as a platform for reproducible research
The other day I was looking at the release notes for the recently published release 1.8 of NumPy, the library that is the basis for most of the Scientific Python ecosystem. As usual, it contains a list of new features and improvements, but also sections such as "dropped support" (for Python 2.4 and 2.5) and "future changes", to be understood as "incompatible changes that you should start to prepare for". Dropping support for old Python releases is understandable: maintaining compatibility and testing it is work that needs to be done by someone, and manpower is notoriously scarce for projects such as NumPy. Many of the announced changes are in the same category: they permit removing old code and thus reduce maintenance effort. Other announced changes have the goal of improving the API, and I suppose they were more controversial than the others, as it is rarely obvious that one API is better than another one.
From the point of view of reproducible research, all these changes are bad news. They mean that libraries and scripts that work today will fail to work with future NumPy releases, in ways that their users, who are usually not the authors, cannot easily understand or fix. Actively maintained libraries will of course be adapted to changes in NumPy, but much, perhaps most, scientific software is not actively maintained. A PhD student doing computational reasearch might well publish his/her software along with the thesis, but then switch subjects, or leave research altogether, and never look at the old code again. There are also specialized libraries developed by small teams who don't have the resources to do as much maintenance as they would like.
Of course NumPy is not the only source of instability in the Python platform. The most visible change in the Python ecosystem is the evolution of Python itself, whose 3.x series is not compatible with the initial Python language. It is difficult to say at this time for how long Python 2.x will be maintained, but it is well possible that much of today's scientific software written in Python will become difficult to run ten years from now.
The problem of scientific publications becoming more and more difficult to use is not specific to computational science. A theoretical physicist trying to read Isaac Newton's works would have a hard time, because the mathematical language of physics has changed considerably over time. Similarly, an experimentalist trying to reproduce Galileo Galilei's experiments would find it hard to follow his descriptions. Neither is a problem in practice, because the insights obtained by Newton and Galilei have been reformulated many times since then and are available in today's language in the form of textbooks. Reading the original works is required only for studying the history of science. However, it typically takes a few decades before specific results are universally recognized as important and enter the perpetually maintained canon of science.
The crucial difference with computations is that computing platforms evolve much faster than scientific research. Researchers in fields such as physics and chemistry routinely consult original research works that are up to thirty years old. But scientific software from thirty years ago is almost certainly unusable today without changes. The state of today's software thirty years from now is likely to be worse, since software complexity has increased significantly. Thirty years ago, the only dependencies a scientific program would have is a compiler and perhaps one of a few widely known numerical libraries. Today, even a simple ten-line Python script has lots of dependencies, most of the indirectly through the Python interpreter.
One popular attitude is to say: Just run old Python packages with old versions of Python, NumPy, etc. This is an option as long as the versions you need are recent enough that they can still be built and installed on a modern computer system. And even then, the practical difficulties of working with parallel installation of multiple versions of several packages are considerable, in spite of tools designed to help with this task (have a look at EasyBuild, hashdist, conda, and Nix or its offshoot Guix).
An additional difficulty is that the installation instructions for a library or script at best mention a minimum version number for dependencies, but not the last version with which they were tested. There is a tacit assumption in the computing world that later versions of a package are compatible with earlier ones, although this is not true in practice, as the example of NumPy shows. The Python platform would be a nicer place if any backwards-incompatible change were accompanied by a change in package name. Dependencies would then be evident, and the different incompatible versions could easily be installed in parallel. Unfortunately this approach is rarely taken, a laudable exception being Pyro, whose latest incarnation is called Pyro4 to distinguish it from its not fully compatible predecessors.
I have been thinking a lot about this issue recently, because it directly impacts my ActivePapers project. ActivePapers solves the dependency versioning problem for all code that lives within the ActivePaper universe, by abandoning the notion of a single collection of "installed packages" and replacing it by explicit references to a specific published version. However, the problem persists for packages that cannot be moved inside the ActivePaper universe, typically because of extension modules written in a compiled language. The most fundamental dependencies of this kind are NumPy and h5py, which are guaranteed to be available in an ActivePapers installation. ActivePapers does record the version numbers of NumPy and h5py (and also HDF5) that were used for each individual computation, but it has currently no way to reproduce that exact environment at a later time. If anyone has a good idea for solving this problem, in a way that the average scientist can handle without becoming a professional systems administrator, please leave a comment!
As I have pointed out in an earlier post, long-term reproducibility in computational science will become possible only if the community adopts a stable code representation, which needs to be situated somewhere in between processor instruction sets and programming languages, since both ends of this spectrum are moving targets. In the meantime, we will have to live with workarounds.
ActivePapers for Python
ActivePapers is a framework for doing and publishing reproducible research. An ActivePaper is a file that contains code (Python modules and scripts) and data (HDF5 datasets), plus the dependency information between all these pieces. You can change a script and re-run all the computations that depend on it, for example. Once your project is finished, you can publish the ActivePaper as supplementary material to your standard paper. You can also re-use code and data from a published ActivePaper by using DOI-based links, although for the moment this works only for ActivePapers stored on Figshare.
I consider this first release of ActivePapers quite usable (I use it, after all), but it's definitely for "early adopters". You should be comfortable working with command-line tools, for example, and of course you need some experience with writing Python scripts if you want to create your own ActivePaper. For inspecting data, you can use any HDF5-based tool, such as HDFView, though this makes sense only for data that generic tools can handle. My first published ActivePaper contains lots of protein structures, which HDFView doesn't understand at all. I expect tool support for ActivePapers to improve significantly in the near future.
Platforms for reproducible research
I'll start with a quote from the manifesto that shows that we have similar ideas of the time scales over which computational research should be reproducible:
"It may be true that code you make available today can be built with only minor pain by many people on current computers. That is unlikely to be true in 5 years, and hardly credible in 20."
So the question is: how can we best ensure that the software used in our computational studies can still be run, with reasonable effort, 20 years from now. To answer that question, we have to look at the possible platforms for computational research.
By a "platform", I mean the combination of hardware and software that is required to use a given piece of digital information. For example, Flash video requires a Flash player and a computer plus operating system that the Flash player can run on. That's what defines the "Flash platform". Likewise, today's "Web platform" (a description that requires a date stamp to be precise, because Web standards evolve so quickly) consists of HTML5, JavaScript, and a couple of related standards. If you want to watch a Flash video in 20 years, you will need a working Flash platform, and if you want to use an archived copy of a 2013 Web site, you need the 2013 Web platform.
If you plan to distribute some piece of digital information with the hope that it will make sense 20 years from now, you must either have confidence in the longevity of the platform, or be willing and able to ensure its long-term maintenance yourself. For the Flash platform, that means confidence in Adobe and its willingness to keep Flash alive (I wouldn't bet on that). For the 2013 Web platform, you may hope that its sheer popularity will motivate someone to keep it alive, but I wouldn't bet on it either. The Web platform is too complex and too ill-defined to be kept alive reliably when no one uses it in daily life any more.
Back to computational science. 20 years ago, most scientific software was written in Fortran 77, often with extensions specific to a machine or compiler. Much software from that era relied on libraries as well, but they were usually written in the same language, so as long as their source code remains available, the platform for all that is a Fortran compiler compatible with the one from back then. For standard Fortran 77, that's not much of a problem, whereas most of the vendor-specific extensions have disappeared since. Much of that 20-year-old software can in fact still be used today. However, reproducing a computational study based on that software is a very different problem: it also requires all the input data and an executable description of the computational protocol. Even in the rare case that all that information is available, it is likely to depend on lots of other software pieces that may not be easy to get hold of any more. The total computational platform for a given research project is in fact as ill-defined as the 2013 Web platform.
Today's situation is worse, because we use more diverse software written in more different languages, and also use more interactive software whose use is notoriously non-reproducible. The only aspect where we have gained in standardization is the underlying hardware and OS layer: pretty much all computational science is done today on x86 processors running Linux. Hence the idea of conserving the full operating environment in the form of a virtual machine. Just fire up VirtualBox (or one of the other virtual machine managers) and run an exact copy of the original study's work environment.
But what is the platform required to run today's virtual machines? It's VirtualBox, or one of its peers. Note however that it's not "any of today's virtual machine managers" because compatibility between their virtual machine formats is not perfect. It may work, or it may not. For simplicity I will use VirtualBox in the following, but you can substitute another name and the basic arguments still hold.
VirtualBox is a highly non-trivial piece of software, and it has very stringent hardware requirements. Those hardware requirements are met by the vast majority of today's computing equipment used in computational science, but the x86 platform is losing market share rapidly on the wider computing device market. VirtualBox doesn't run on an iPad, for example, and probably it never will. Is VirtualBox likely to be around in 20 years? I won't dare a prediction. If x86 survives for another 20 years AND if Oracle sees a continuing interest in this product, then it will. I won't bet on it though.
What we really need for long-term recomputability is a simple platform. A platform that is simple enough that the scientific community alone can afford to keep it alive for its own needs, even if no one else in the world cares about it.
Unfortunately there is no suitable platform today, to the best of my knowledge. Which is why virtual machines are perhaps the best option right now, for lack of a satisfactory one. But if we care about recomputability, we should design and develop a good supporting platform, starting as soon as possible.
For a more detailed discussion of this issue, see this paper written by yours truly. It comes to the conclusion that the closest existing approximation to a good platform is the Java virtual machine. What we'd want ideally is something similar to the JVM, but designed and optimized for scientific applications. A basic JVM implementation is quite simple (the complex JIT stuff is not a requirement), a few orders of magnitude simpler than VirtualBox, and it has no specific hardware dependencies. It's even simpler than many of today's scientific software packages, so the scientific community can definitely afford to keep it alive, The tough part is... no, it's not designing or writing the required software, it's agreeing on a specification. Perhaps it will never happen. Perhaps virtual machines will remain the best choice for lack of a satisfactory one. Or perhaps we will end up compiling our software to asm.js and run in the browser, just because someone else will keep that platform alive for us, no matter how ill-adapted it is to our needs. But don't say you haven't been warned.
Bye bye Address Book, welcome BBDB
What started to annoy me seriously about Address Book is a bug that makes the database and its backups grow over time, even if no contacts are added, because the images for the contacts keep getting copied and never deleted under certain circumstances. I ended up having address book backups of 200 MB for just 500 contacts, which is ridiculous. A quick Web search shows that the problem has been known for years but has not yet been fixed.
When I upgraded from MacOS 10.6 to 10.7 about a year ago (I am certainly not an early adopter of new MacOS versions), I had a second reason to dislike Address Book: the user interface had been completely re-designed and become a mess in the process. Every time I use it I have to figure out again how to navigate groups and contacts.
I had been considering moving to BBDB for a while, but I hadn't found any good solution for synchronizing contacts with my Android phone. That changed when I discovered ASynK, which does a bi-directional synchronization between a BBDB database and a Google Contacts account. That setup actually works better than anything I ever tried to synchronize Address Book with Google Contacts, so I gained more than I expected in the transition.
At first glance, it may seem weird to move from technology of the 2000's to technology of the 1970's. But the progress over that period in managing rather simple data such as contact information has been negligible. The big advantage of the Emacs platform over the MacOS platform is that it doesn't try to take control over my data. A BBDB database is just a plain text file whose structure is apparent after five minutes of study, whereas an Address Book database is stored in a proprietary format. A second advantage is that the Emacs developer community fixes bugs a lot faster than Apple does. A less shiny (but perfectly usable) user interface is a small price to pay.
A critical view of altmetrics
At first sight, altmetrics appear as an evident "update" to traditional bibliometry. It sounds pretty obvious that, as scientific communication moves on to new media and finds new forms of expressions, bibliometry should adapt. On the other hand, bibliometry is considered a more less necessary evil by most scientists. Many deplore today's "publish or perish" culture and correctly observe that it is harmful to science in the long term, giving more importance to the marketing of research studies than to their careful design and meticulous execution. I haven't yet seen any discussion of this aspect in the context of altmetrics, so I'd like to start such a discussion with this post.
First of all, why is bibliometry so popular, and why is it harmful in the long run? Second, how will this change if and when altmetrics are adopted by the scientific community?
Bibliometry provides measures of scientific activity that have two important advantages: they are objective, based on data that anyone can check in principle, and they can be evaluated by anyone, even by a computer, without any need to understand the contents of scientific papers. On the downside, those measures can only indirectly represent scientific quality precisely because they ignore the contents. Bibliometry makes the fundamental assumption that the way specific articles are received by the scientific community can be used as a proxy for quality. That assumption is, of course, wrong, and that's how bibliometry ultimately harms the progress of science.
The techniques that people use to improve their bibliometrical scores without contributing to scientific progress are well known: dilution of content (more articles with less content per article), dilution of authorship (agreements between scientists to add each others' names to their works), marketing campaigns for getting more citations, application of a single technique to lots of very similar applications even if that adds no insight whatsoever. Altmetrics will cause the same techniques to be applied to datasets and software. For example, I expect scientific software developers to take Open Source libraries and re-publish them with small modifications under a new name, in order to have their name attached to them. Unless we come up with better techniques for software installation and deployment, this will probably make the management of scientific software a bit more complicated because we will have to deal with lots of small libraries. That's a technical problem that can and should be solved with a technical solution.
However, these most direct and most discussed negative consequences of bibliometry are not the only ones and perhaps not the worst. The replacement of expert judgement by majority vote, which is the basis of bibliometry, also in its altmetrics incarnation, leads to a phenomenon which I will call "scientiic bubbles" in analogy to market bubbles in economy. A market bubble occurs if the price of a good is determined not by the people who buy it to satisfy some need, but by traders and speculators who try to estimate the future price of the good and make a profit from a rise or fall relative to the current price. In science, the "client" whose "need" is fulfilled by a scientific study is mainly future science, plus in the case of applied research engineering and product development. The role of traders and speculators is taken by referees and journal editors. A scientific bubble is a fashionable topic that many people work on not because of its scientific interest but because of the chance it provides to get a highly visible publication. Like market bubbles, scientific bubbles eventually explode when people realize that the once fashionable topic was a dead end. But before exploding, a bubble has wasted much money and intellectual energy. It may also have blocked alternative and ultimately more fruitful research projects that were refused funding because they were in contradiction with the dominating fashionable point of view.
My prediction is that altmetrics will make bubbles more numerous and more severe. One reason is the wider basis of sources from which references are counted. In today's citation-based bibliometry, citations come from articles that went through some journal's peer-reviewing process. No matter how imperfect peer review is, it does sort out most of the unfounded and obviously wrong contributions. To get a paper published in a journal whose citations count, you need a minimum of scientific competence. In contrast, anyone can publish an opinion on Twitter or Facebook. Since for any given topic the number of experts is much smaller than the number of people with just some interest, a wider basis for judgement automatically means less competence on average. As a consequence, high altmetrics scores are best obtained by writing articles that appeal to the masses who can understand what the work is about but not judge if it is well-founded. Another reason why altmetrics will contribute to bubbles is the positive feedback loop created by people reading and citing publications because they are already widely read and cited. That effect is dampened in traditional bibliometry because of the slowness of the publishing and citation mechanism.
My main argument ends here, but I will try to anticipate some criticisms and reply to them immediately.
One objection I expect is that the analysis of citation graphs can be used to assign a kind of reputation to each source and weight references by this reputation. That is the principle of Google's famous PageRank algorithm. However, any analysis of the citation graph suffers from the same fundamental problem as bibliometry itself: a method that only looks at relations between publications but not at their contents can't distinguish a gem from a shiny bubble. There will be reputation bubbles just like there are topic bubbles. No purely quantitative analysis can ever make a statement about quality. The situation is similar to mathematical formalisms, with citation graph analysis taking the role of formal proof and scientific quality the role of truth in Gödel's incompleteness theorem.
Another likely criticism is that the concept of the scientific bubble is dubious. Many paths of scientific explorations have turned out to be failures, but no one could possibly have predicted this in the beginning. In fact, many ultimately successful strategies have initially been criticized as hopeless. Moreover, exploration of a wrong path can still lead to scientific progress, once the mistake has been understood. How can one distinguish promising but ultimately wrong ideas from bubbles? The borderline is indeed fuzzy, but that doesn't mean that the concept of a bubble is useless. It's the same for market bubbles, which exist but are less severe when a good is traded both for consumption and for speculation. My point is that the bubble phenomenon exists and is detrimental to scientific progress.
Lessons from sixteen years of molecular simulation in Python
First, a bit of history. The first version of MMTK was released about 16 years ago. I don't have the exact data, but the first message on the MMTK mailing list, announcing MMTK release 1.0b2, is dated 29 May 1997. Back then Python 1.4 was the state of the art and Numerical Python was a young project that was just beginning to stabilize. MMTK was one of the first domain-specific scientific libraries written in Python, at a time when the scientific Python community was very small and its members were mostly considered cranks by their peers. MMTK was designed from the start as a Python library, with relatively small bits of C code for the time-critical stuff (mainly energy evaluation and MD integration), with NumPy arrays at the Python-C interface. This has since become one of the two main approaches to using Python in scientific computing, the other one being wrapper code around libraries written in C/C++ or Fortran.
So what would I do differently if I were to start writing MMTK today? Many things, for different reasons. Lets first get the obvious stuff out of the way: the Python ecosystem has evolved significantly since 1997, and of course I would use Python 3, and Cython instead of C for the time-critical parts. I would also adopt many of the conventions that the community has developed but which weren't around in 1997. I might even be tempted to use bleeding-edge tools like Numba, although with hesitation: Numba is not only a moving target at this time, but also requires dependencies (I am thinking mostly of LLVM) which are big and non-trivial to install. One lesson I have learned in 16 years of scientific Python is that dependencies can cause more trouble than they are worth. It's nice in theory to re-use existing tested code, but it also makes installation and deployment more cumbersome.
So far for changes in the Python ecosystem. What has changed as well, though at a slower pace, is the role of computation in science and in particular in molecular simulation. Back in 1997, there were a few molecular simulation ecosystems that operated almost in isolation. The big players were the CHARMM, AMBER, and GROMOS/GROMACS communities. Each of them had their own software, their own file formats, and their own force fields. Members of these communities would of course talk about science to each other, but not share any software or data. Developing new computational methods required a serious investment into one of these ecosystems. That was in fact my main motivation for developing MMTK: I figured that I would be more efficient (not to mention more satisfied) writing a new system from scratch using modern development tools than trying to get familiar with crufty Fortran code. But I adopted basically the same approach with MMTK: I created a new ecosystem without much regard to sharing code or data with the rest of the world. As an illustration, MMTK defines its own trajectory format which I still consider superior to what the rest of the world is doing, but which is undeniably hard to use without MMTK, given that the definition of a universe is stored as an executable Python expression. MMTK also encourages storing data as Python pickle files, which are even harder to deal with for other programs.
Today we are seeing a change in attitude in computational science that I am sure will soon reach the molecular simulation community as well. People are starting to realize that computational results have serious reliability problems. The most publicized case in the structural biology community was the retraction of a few important published protein structures following the discovery of a bug in the data processing software that lead to completely wrong final structures. This and similar events point to the urgent need for better validation of computational results. One aspect of validation is re-running the same computation with different tools. Another aspect is publishing both software and raw data, enabling other scientists to inspect them and check their validity. Technology for sharing scientific code and data exists today (have a look at Github, Bitbucket, and figshare, for example). But in molecular simulation, there are still important practical barriers to such validation attempts, in particular the use of program-specific and badly documented file formats. While MMTK's file formats are documented, they are still program-specific and thus incompatible with the requirements of the future.
The sentence that I would like to write now is "If I were to rewrite MMTK today, I would use the exchange data formats accepted by the molecular simulation community". But those formats don't exist yet, although there are a few initiatives to develop them. My own contribution to this effort is the Mosaic data model and data formats - if you are interested in this subject, please have a look at it and send me your feedback. Mosaic will of course find its way into future versions of MMTK.
Finally, there are things I would do differently because the experience with MMTK has shown that a few initial design decisions were not the best ones. Number one is the absence of stable atom numbers. In MMTK, each atom and molecule is represented by a unique Python object, and there are ways to refer uniquely to everything by using Python expressions. But there is no such thing as a unique order of atoms that would assign a number to each one. Atoms do have numbers by which the low-level C code refers to them, but these numbers can be different every time you run a Python script. My original design goal was to discourage the use of numbers to refer to atoms, because this is an important source of mistakes if the simulated system undergoes changes. But every other molecular simulation program out there uses numbers to refer to atoms, so people are used to them. For interoperability with other programs, atom numbers are fundamental. There are ways to handle such situations, of course, but it's a constant source of headaches.
The other design aspect that I would change if I were to rewrite MMTK today is the hierarchy of chemical objects. MMTK has Atoms, Groups, Molecules, and Complexes, plus specializations such as AminoAcidResidue (a special Group), PeptideChain (a special Molecule), and Protein (a special Complex). While all of these correspond to some chemical reality, the system is more complex than required for molecular simulation, leading in some situations to code that is bloated by irrelevant special cases. Today I'd go for just Atoms and Groups, with special features of specific kinds of groups indicated by attributes rather than specific classes.
Integrating scientific software and datasets into the citation record
This morning I read C. Titus Brown's blog post on how science could be so much better if scientitic data and the software used to work with it were openly available for reuse. One problem he mentions, like many others have done before, is the lack of incentive for publishing anything else but standard scientific papers. What matters for a scientist's career and for grant applications is papers, papers, papers. Any contribution that's not in a scientific journal with a reputation and an impact factor is usually ignored, even if its real impact exceeds that of many papers that nobody really wants to read.
Ideally, published scientific data and software should be treated just like a paper: it should be citeable and it should appear in the citation databases that are used to calculate impact factors, h factors, and whatever other metrics bibliometrists come up with and evaluation committees appreciate for their ease of use.
Treating text (i.e. papers), data, and code identically also happens to be useful for making scientific publications more useful to the reader, by adding interactive visualization and exploration of procedures (such as varying parameters) to the static presentation of results in a standard paper. This idea of "executable papers" has generated a lot of interest recently, as shown by Elsevier's Executable Paper Challenge and the Beyond the PDF workshop. For a technical description of how this can be achieved, see my ActivePapers project and/or the paper describing it. In the ActivePapers framework, a reference to code being called, or to a dataset being reused, is exactly identical to a reference to a published paper. It would then be much easier for citation databases to include all references rather than filter out the ones that are "classical" citations. And that's a good motivation to finally treat all scientific contributions equally.
Since the executable papers idea is much easier to sell than the idea of an upated incentive system, a seemingly innocent choice in technology could end up helping to change the way scientists and research projects are evaluated.
Tags: computational science, computer-aided research, emacs, mmtk, mobile computing, polycrisis, programming, proteins, python, rants, reproducible research, science, scientific computing, scientific software, social networks, software, source code repositories, sustainable software
By month: 2026-03, 2025-11, 2025-06, 2025-04, 2025-03, 2024-10, 2023-11, 2023-10, 2022-08, 2021-06, 2021-01, 2020-12, 2020-11, 2020-07, 2020-05, 2020-04, 2020-02, 2019-12, 2019-11, 2019-10, 2019-05, 2019-04, 2019-02, 2018-12, 2018-10, 2018-07, 2018-05, 2018-04, 2018-03, 2017-12, 2017-11, 2017-09, 2017-05, 2017-04, 2017-01, 2016-05, 2016-03, 2016-01, 2015-12, 2015-11, 2015-09, 2015-07, 2015-06, 2015-04, 2015-01, 2014-12, 2014-09, 2014-08, 2014-07, 2014-05, 2014-01, 2013-11, 2013-09, 2013-08, 2013-06, 2013-05, 2013-04, 2012-11, 2012-09, 2012-05, 2012-04, 2012-03, 2012-02, 2011-11, 2011-08, 2011-06, 2011-05, 2011-01, 2010-07, 2010-01, 2009-09, 2009-08, 2009-06, 2009-05, 2009-04